(CUSTOM) Protein Microarray
Protein microarrays
With its high density protein microarray technology, Immunostep offers its clients a high performance tool for the identification of biomarkers, the identification of enzymatic substrates, profiles of antibody specificity, auto-antibody tests, pharmacological targets and studies of protein-protein interaction. For the customised development of protein microarrays, the organisation has a cDNA library or gene library, with more than 10,000 clones of different human proteins, amongst which the presence of over 500 kinase clones, a complete representation of the Human Kinome, should be noted. This means that we can offer personalised fabrication of high density protein microarrays, with hundreds or thousands of diverse human proteins, from the expression of the cDNA library, including kinases, phosphatases, nuclear receptors and receptors coupled to a G-protein, among others. These proteins are immobilised with multiple replicas, depending on the client’s request, using a previously functionalised glass slide as a solid substrate.

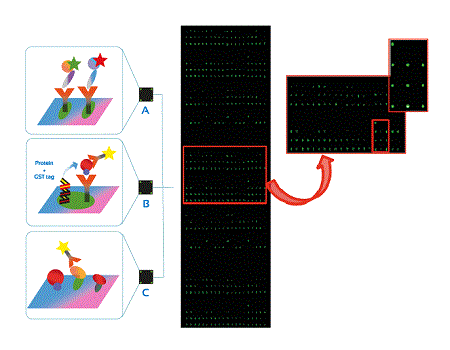
Figure 1: Different microarray formats A) of antibodies B) tagged proteins C) proteins and array detection assay of proteins scanned by utilising Cy3 adjustments.
Protein microarray technology allows us to obtain results in a very short period of time, typically a one-day assay, in a straightforward way, with very little sample volume and on a large scale or with high yield.
Flow Chart Diagram for the Customised Development of a Protein Microarray:


Figure 2: Robotic platform for the preparation of printing plates, incubation and revelation of protein arrays (EVO 150, Tecan – Immunostep facilities).
Massive Screening of Phage Display in Array Format
Emerging technologies in the design and generation of antibodies require improved approaches to their detection, characterisation and validation. More specifically, “Phage Display” technology constitutes a powerful tool for the generation of specific antibodies against certain types of antigens, given the smallness of the single chain variable fragments (scFvs) and their ability to penetrate tissues.
This technology benefits from various others developed by Immunostep. In this sense, we offer a platform for the screening of thousands of clones simultaneously, with a reproducibility and sensitivity superior to the more usual ELISA techniques. It is an innovative platform which provides an effective yet simple methodology that permits the identification of scFv. The platform is based on the use of a nitrocellulose array which enables us to immobilise thousands of clones and later carry out their evaluation with fluorescence techniques against the desired antigen, combining this with a bioinformatic tool and an algorithm that allows us to compare the results obtained with those collected using conventional screening techniques.

Figure 3: General overview for the construction, selection and analysis of phage-display libraries using ELISA and microarray assays. After the construction of the library (1), a selection is made via washing cycles, elution and amplification (2). ELISA and microarray technology have been used to analyse the selected phages against specific antigens (3).
Identification of clones or positive phages: we take into account the values of fluorescence intensity at wavelengths of 532 nm obtained by analysing each array in a fluorescence scanner.

Fig. 4 General view of the analysis of phage display libraries through microarrays. Incubation of positive clones using fluorescently conjugated antigen (1), revelation after incubation by means of a fluorescence scanner (2), analysis of the results using a bioinformatic tool (3).
Flow Cytometry- From Research to Clinical Analysis
Flow Cytometry is a technology used in the analysis of particles and/or cells in suspension. It permits the multiparametric and simultaneous analysis of physical and chemical characteristics at a speed of thousands of cells per second. Flow cytometry applications are very varied and range from basic to clinical research.
At Immunostep, we offer a wide battery of optimised cell assays that allow us to obtain results rapidly and economically in such a way as to boost our clients’ research.

Fig. 7 Separating Cytometer activated by fluorescence (FACSAria II- Immunostep facilities)
From amongst our cell analysis services, we would like to point out the following:
In vitro drug screening
Permits the evaluation of toxicity and the apoptotic pathways induced by drugs, as well as the screening of hundreds of drugs, enabling us to identify the best treatment or combination of available drugs for each kind of pathology or patient.
Cell Immunophenotyping
Utilising a monoclonal antibody panel, it allows us to evaluate the expression of both membrane and cytoplasmic antigens, permitting the characterisation and classification of different cell populations.
Functional assays
(phagocytosis/oxidative burst): it allows us to determine the function of the phagocytosis of leukocytes (neutrophils and monocytes) in whole blood by measuring the global percentage of phagocytosis of monocytes and granulocytes, and the individual phagocytic activity of each cell. Additionally, it also enables us to determine quantatively the respiratory or oxidative burst that destroys engulfed micro-organisms producing the phagolysosome.

Fig. 5 Analysis of Phagocytosis and Toxicity in the Biological Characterisation of Nanomaterials
Activation, proliferation and cell death
It permits the analysis of activation markers such as IL-2R, L-selectin, CD69 and TNF-α, among others. It is interesting to point out the studies of platelet activation (P-selectin), or of basophils (mCD63) as a response to type 1 hypersensitivity reactions.

Fig. 6 Histogram of lysed normal whole blood sample selected with CD123+/CD203c+/HLA-DR to detect the basophil population versus the population of CD63 + activated basophils.
Dendritic cell (DC)-based assays
Provide an in vitro comparison of the antigenicity of proteins at cell level. The assay is based on the protein or antigen presentation by means of DCs, which facilitate the determination of the relative antigenicity of the diverse candidate proteins, allowing a direct comparison. Donor PBMCs are employed in the assay as a source of monocytes, which are cultivated so as to generate immature DCs. The DCs are charged with the antigen of interest to later induce phenotypic maturing, through cultures in defined media. CD8+ T cells become depleted via magnetic separation using the PBMC of donors of the same sample from which the monocytes were obtained. Afterwards, the PBMCs depleted of CD8+ T cells are labelled with CFSE in order to carry out the analysis and monitoring of the cell division. The PBMC sample labelled with CFSE will subsequently be cultured with the DCs charged with the antigens for seven days. Finally, the culture is evaluated by means of flow cytometry. The assessment will also include antigen controls, using those which are well-known for their high antigenicity.
How Can We Help You?
Immunostep’s service team has long and valuable experience in the development and utilisation of protein arrays, as well as in the development of customised assays in which flow cytometry is used as a platform. Both are powerful tools with many possible applications in research and in clinical practice. Below are examples of some of the services we have carried out:
| Nº | Service | Description |
| 1 | Antibody array | Antibody array for simultaneous determination of 70 different human leukocyte antigens in peripheral blood mononuclear cells. |
| 2 | Screening and selection of scFv | Screening and selection by means of phage microarray (scFv) derived from Tomlinson I+J library, IL8 specific. |
| 3 | Cell Immunophenotyping | Multiparametric analysis of cytokines (INFγ, IL-2, TNFα) at intracellular level in stimulated T cells. |
| 4 | Functional assays | Phagocytosis analysis; cell activation and death in the characterisation of superparamagenetic nano-particles in a customised assay to carry out the analysis in a single tube. |
| 5 | Cell activation | Evaluation of basophil activation after incubation with a battery of allergens. |
| 6 | Cell assays of DCs | Evaluation of the antigenicity of proteins derived from Mycobacterium tuberculosis, as potential candidates for vaccine development. |





